In the medical field , particularly in cancer research, there is a growing emphasis on using machine learning to detect and analyze diseases through image analysis. Histopathology image classification, especially involving images stained with Hematoxylin and Eosin (H&E), is becoming increasingly crucial for automated detection of complex tissue structure and cancer-grade classification. Many existing single and hybrid models either involve complex pre-processing steps or concatenate the outputs of two or more models, which results in complexity and inefficiency. In contrast, our proposed ‘‘TransNetV’’ model utilizes the local feature extraction capabilities of CNNs and then serially passes these features through the global contextual understanding of the Transformer’s attention mechanism. This methodology leverages the weight-sharing properties of CNNs and the ability of Transformers to comprehend the broader context of spatial relationships in large-scale patterns, making it effective on datasets of varying sizes and complex feature analyses. For the experiment, the model is first trained on the National Center for Tumor Diseases (NCT Biobank) CRC dataset and is then tested on the CRC-HE-VAL-7K external dataset. To further investigate our model’s performance and generalization ability,it is also trained and tested on the Kather_texture_2016_5000 dataset. Both validation sets CRC-HEVAL- 7k and Kather_texture_2016_5000 datasets achieved exceptional classification accuracies of 98.54% and 98.96%, respectively. Furthermore, the proposed TransNetV architecture is adaptable to other convolutional models such as ResNet50, ResNet101, and VGG19. The results of our methodology surpass many existing state-of-the-art solutions found in the literature.
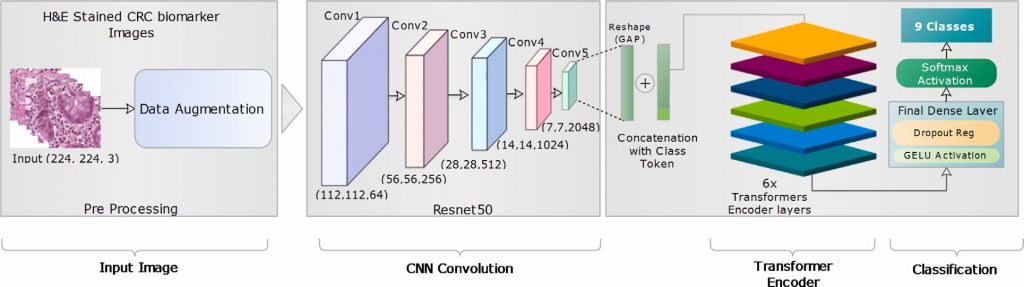
The proposed framework introduces a unified model that combines the abilities of complex feature recognition of the Convolutional Neural Networks with the spatial understanding of the Transformer architecture.
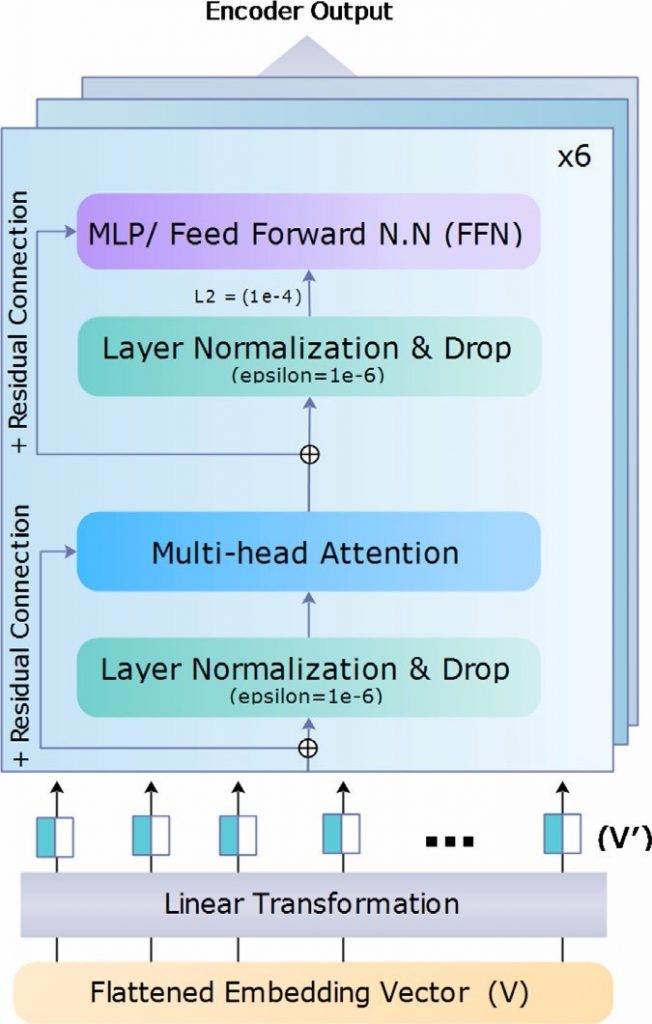
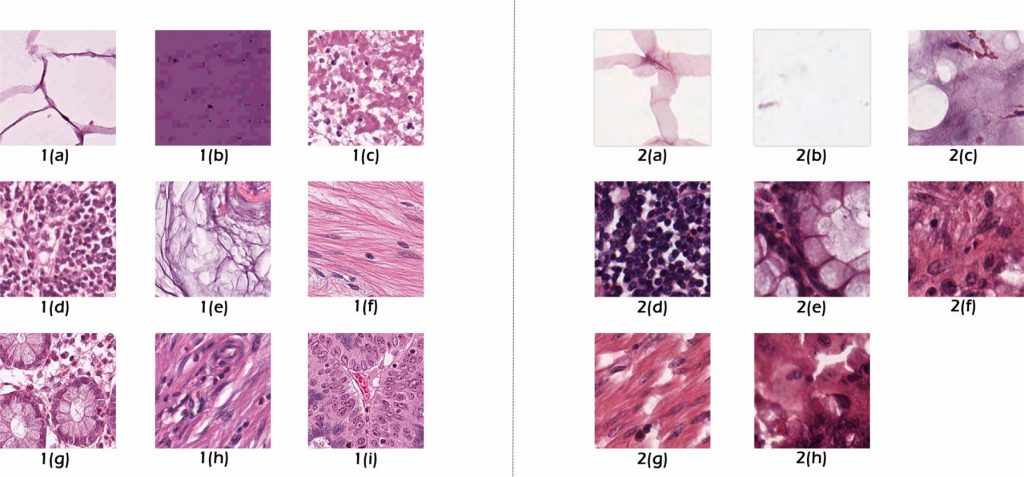
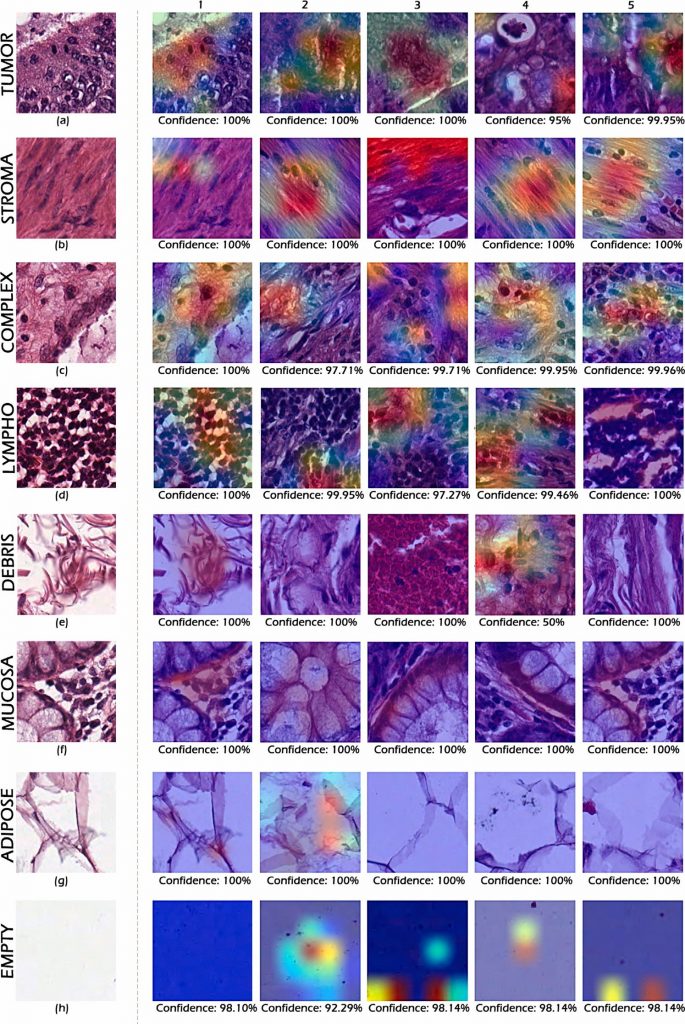
Relevant Publicatoins
- Tanveer, Mohsan, Muhammad Usman Akram, and Asad Mansoor Khan. “TransNetV: An optimized hybrid model for enhanced colorectal cancer image classification.” Biomedical Signal Processing and Control 96 (2024): 106579.