At BIOMISA, we are doing multi-disciplinary research. Our current research work includes:
Deep learning applications
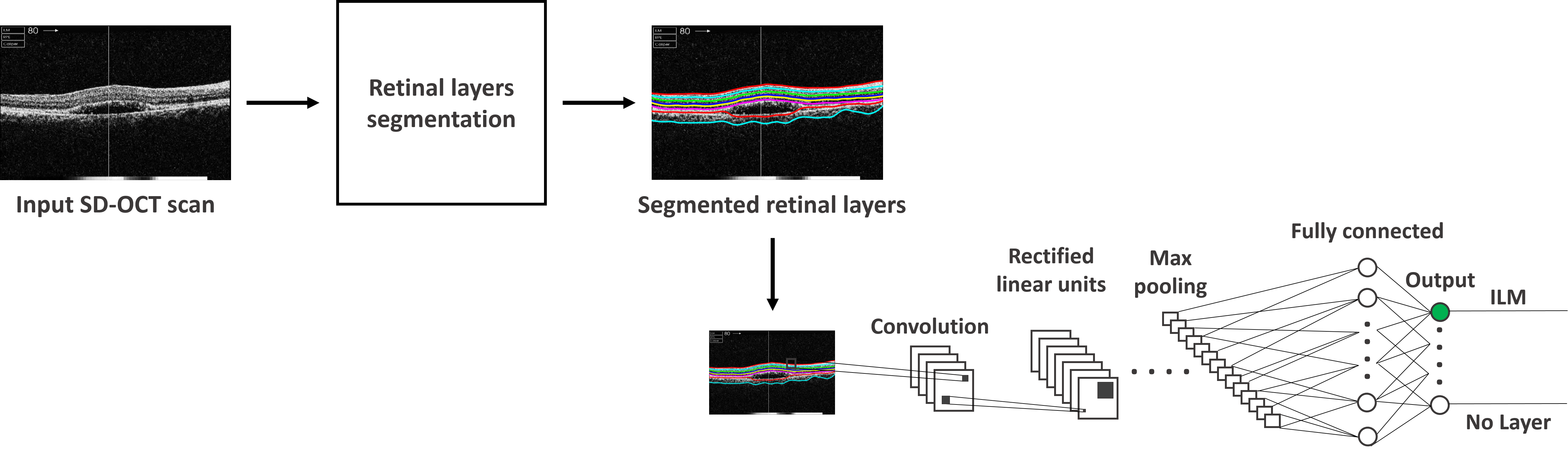
Deep learning for OCT image analysis:
Extraction of retinal layers from optical coherence tomography (OCT) scans is critical for analyzing retinal anomalies and manual segmentation of these retinal layers is a very cumbersome task. Recently, deep learning has gained much popularity in medical image analysis due to its underlying precision and robustness. We at BIOMISA have developed a deep convolutional neural network and structure tensor-based segmentation framework (CNN-STSF) for the fully automated segmentation of up to eight intra-retinal layers from normal as well as diseased OCT scans. First of all, the proposed framework computes coherent tensor from the candidate scan through which retinal layers are extracted. Afterwards, the pixels representing the layers are further classified using cloud-based deep convolutional neural network (CNN) model trained on 1,200 retinal layers patches. CNN model in the proposed framework computes the probability of each layer pixels and assign it to be part of that layer for which it has the highest probability. The proposed framework has been tested and validated on more than 35,000 OCT scans from different publicly available datasets and from local Armed Forces Institute of Ophthalmology (AFIO) dataset.
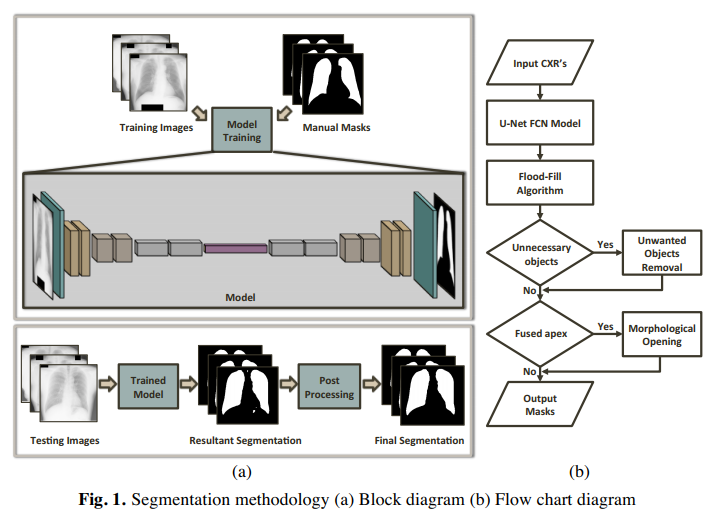
Deep learning for chest x-rays images:
Deep neural networks have entirely dominated the machine vision space in the past few years due to their astonishing human comparable performance. Here at BIOMISA, we apply the power of such networks to segment out lungs from chest x-rays, which is a crucial step in any computer-aided diagnostic (CAD) system design in radiology. A fully convolutional network was utilized to extract lungs region from the x-rays and this process is repeated ten times, with random split of data into a 60:40 ratio as training and testing sets respectively, to calculate the average accuracy. The methodology was tested on three datasets: Japanese Society of Radiological Technology (JSRT), Montgomery County (MC), and a local dataset where the propsoed framework achieved mean accuracy of 97.1%, 97.7% & 94.2% respectively. The results proved that the proposed methodology is efficient enough and can be generalized for other such segmentation problems in medical imaging domain.
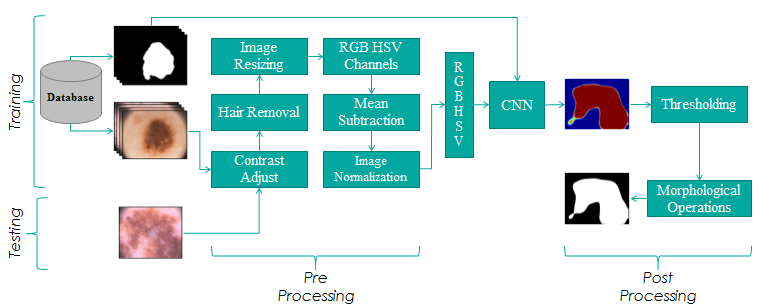
Deep learning for the extraction of skin lesions from dermoscopic images:
We at BIOMISA utilized deep learning for the extraction of skin lesions from dermoscopic images. Our segmentation framework is based on 31 layers which achieve high accuracy for the segmentation of skin lesions. The proposed framework was validated on two publically available databases of PH2 and ISIC 2017 where it achieved the dice coefficient of 92.3% for PH2 Dataset and the dice coefficient of 85.5% for ISIC 2017 dataset.